Happy to announce the 2024 QBioS PhD @ GT Hands on Modeling Workshop! Learn to use AI (AlphaFold2) to examine protein structure and folding! Sign up here:
sites.gatech.edu/qbios-workshop…
Due to budget cuts, this may be the last public workshop we run- sign up soon before it fill up!







Deep learning on computational biology and bioinformatics tutorial: from DNA to protein folding and alphafold2 | AI Summer hubs.ly/Q026lq9z0 #MachineLearning #DeepLearning #ai #ai summer #machinelearning #artificialintelligence #python

👇 from a fantastic, wise letter to Nature by Jennifer Listgarten
“DeepMind researchers performed a fantastic feat with the development of AlphaFold2, substantially moving the needle on the protein structure prediction problem. Protein structure prediction is a tremendously important…

Don't miss 'AI enthusiasm', my blog series posted on the awesome DEV community DEV Community
I talk about the impact of AI on everyday life: my last post is on AlphaFold2, by Google DeepMind, a game-changer in protein science!
dev.to/astrabert/ai-e…
#ai #blog #TECH4ALL


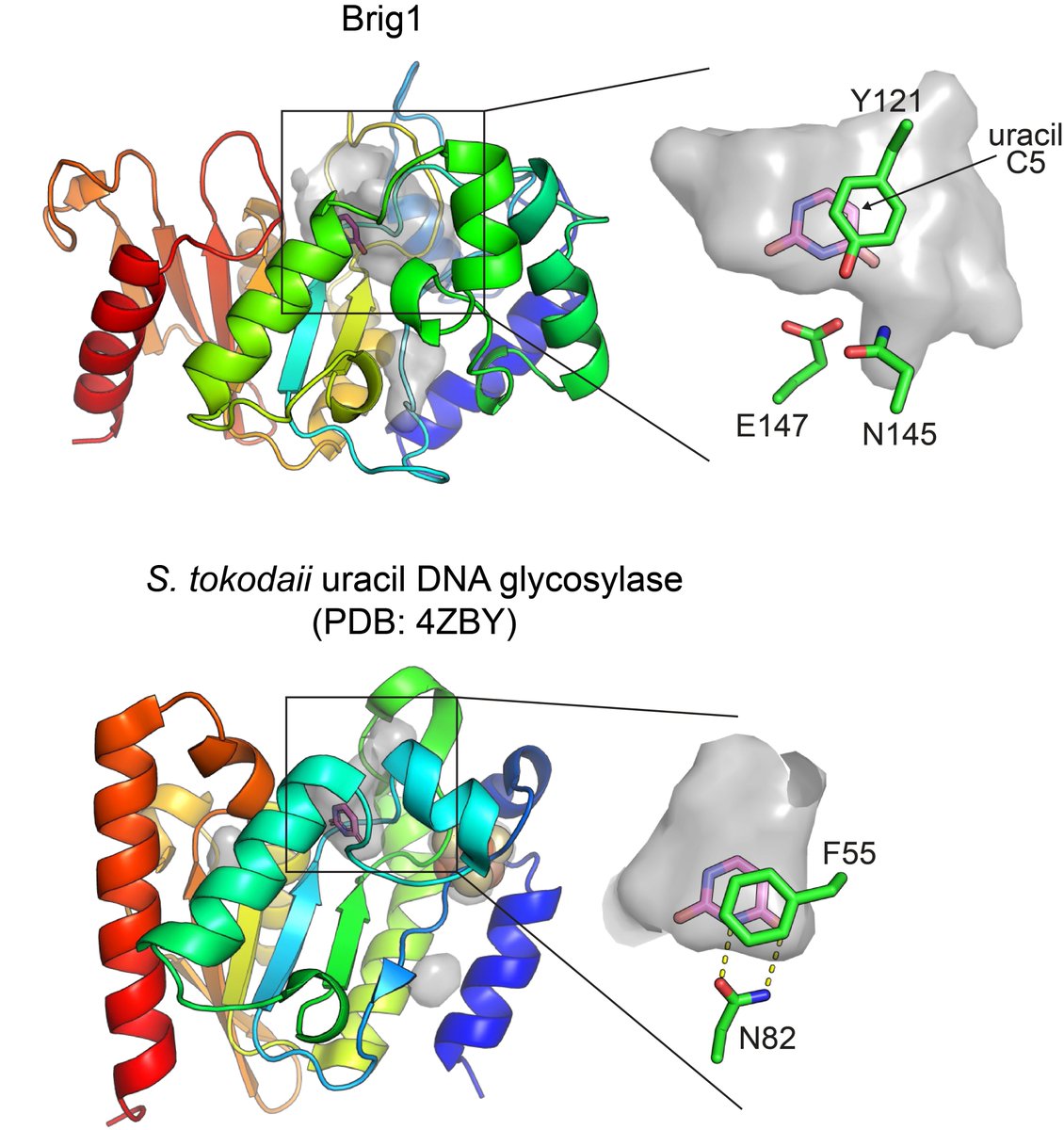
🙌 Happy to share Muhammad Asif Ali and Gustavo Caetano-Anollés's latest paper:
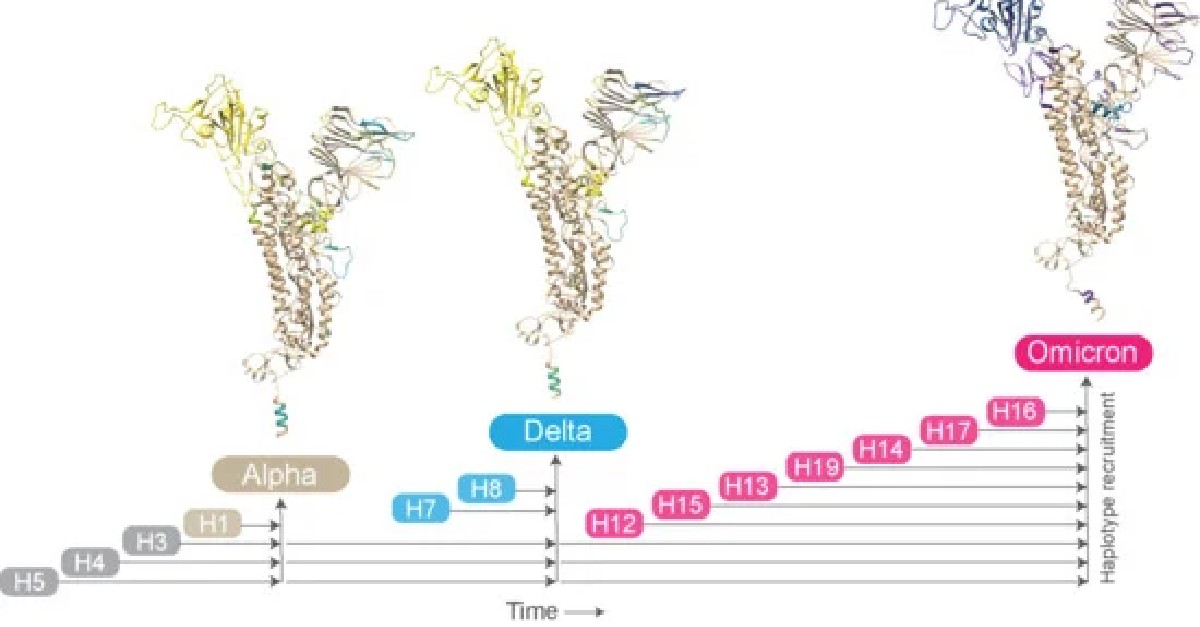
📰 'AlphaFold2 Reveals Structural Patterns of Seasonal Haplotype Diversification in SARS-CoV-2 Spike Protein Variants'
🔦 Full text at brnw.ch/21wIT7D.
University of Illinois




torusengoku💙💛 3.5 Å 以上くらいのデータ・5 Å くらいまでのデータ・それ未満の低分解能データ(例えば、7 Å に AlphaFold2 のモデルを rigid body fit したようなもの)では、含まれる情報が違いすぎるし、登録方法も検証方法も異なるべきだと思うのですが、今は一括りなのがとりとしてはつらいです。

Amy Lu Gave a course at Heidelberg about structural bioinformatics last academic year including an AF2 lecture: structural-bioinformatics.netlify.app
Heavily built upon great resources already mentioned such as Nazim Bouatta's lectures. Best post for me is still this one: moalquraishi.wordpress.com/2021/07/25/the…



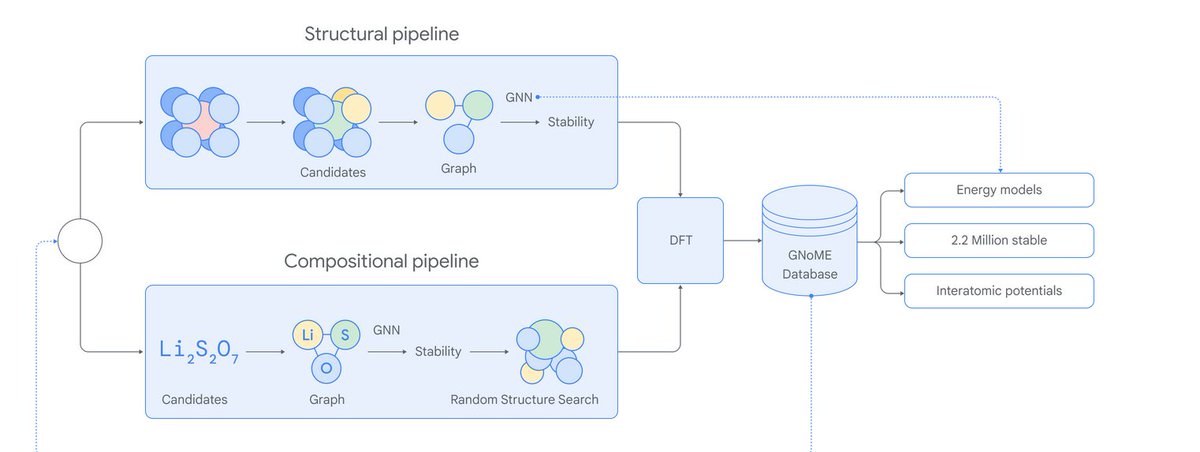
![Sanchit Misra (I am hiring) (@sanchit_misra) on Twitter photo 2024-04-11 18:31:34 The protein folding pipeline is AlphaFold2-based and consists of database search, multiple sequence alignment and AlphaFold2 evoformer and structure modules. For a set of proteins of length < 1K, Open Omics gets 17x speedup vs. prior CPU baseline on the end-to-end time. [3/n] The protein folding pipeline is AlphaFold2-based and consists of database search, multiple sequence alignment and AlphaFold2 evoformer and structure modules. For a set of proteins of length < 1K, Open Omics gets 17x speedup vs. prior CPU baseline on the end-to-end time. [3/n]](https://pbs.twimg.com/media/GK524VmWcAEfS2N.jpg)