
Fantastic collaboration with Big Data Biology Lab. With machine learning, we uncovered a whole new world of antimicrobials (~1 M!) in this first exploration of the entire global microbiome + provide ground-truth experimental validation in preclinical models.
biorxiv.org/content/10.110…

I have had many undergraduate students recently ask me about computational modelling in biology. It’s fast becoming more popular and accessible. Enthusiasm for the subject makes me really happy 👩🏼💻🧬🦠 we happen to have a great masters course Cardiff University School of Biosciences #bigdatabiology

Quarterly updates from our lab
- We focus on Vedanth, an #EmergentVentures visitor from India, working on argNorm (normalizing antibiotic resistance gene annotations)
- Several publications & tool updates since the last update!
- Open office hours in April with Luis


Big Data Biology Lab @EMBARK_JPIAMR Greetings from Trento to Belgrade!
Didn’t miss opportunity for Microbiome Virtual International Forum invitation ;)
📸 Milka

.Luis Pedro Coelho🌻🇮🇱 presents the metagenomic binner SemiBin2. It applies self-supervised contrastive learning for separation, is fast and showing robust performance for known and novel species. #ISMBECCB2023
📄 doi.org/10.1093/bioinf…
Code: github.com/BigDataBiology…



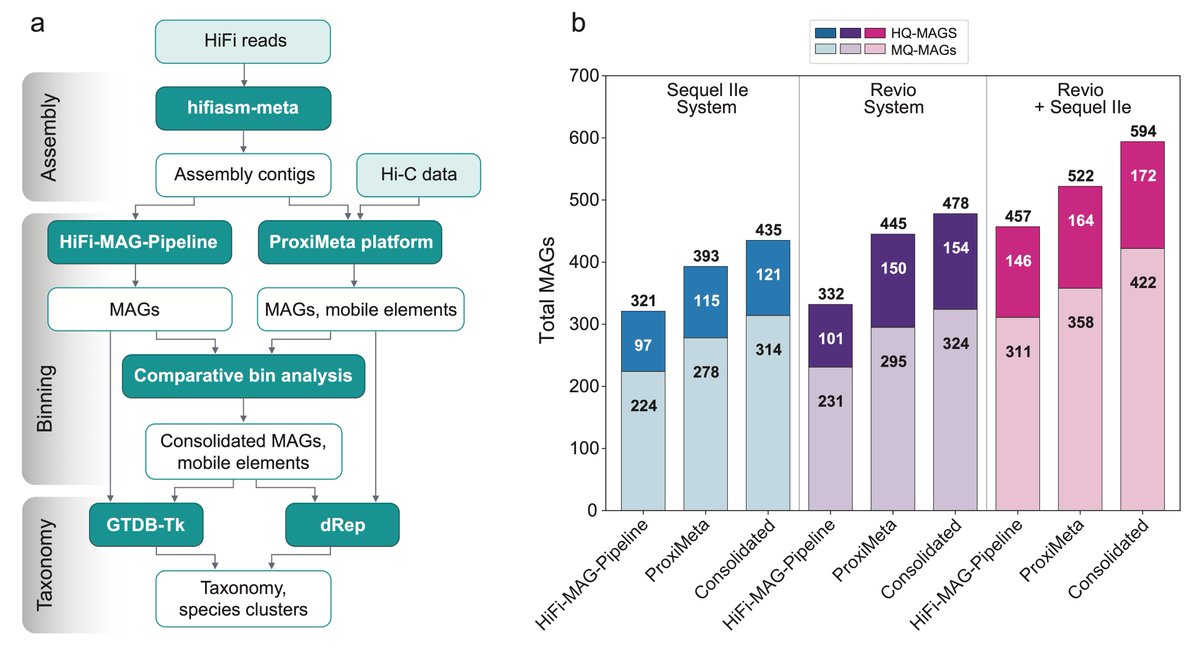
Florian Plaza Oñate Luis Pedro Coelho🌻🇮🇱 Big Data Biology Lab I think we still need to look at samples of different complexity, and at different depths🤔. Each method (bioinformatic, Hi-C) also picks up unique MAGs missed by the other.
Here is a newer figure for the Zymo results, showing bioinformatic and Hi-C results, plus the merged set.

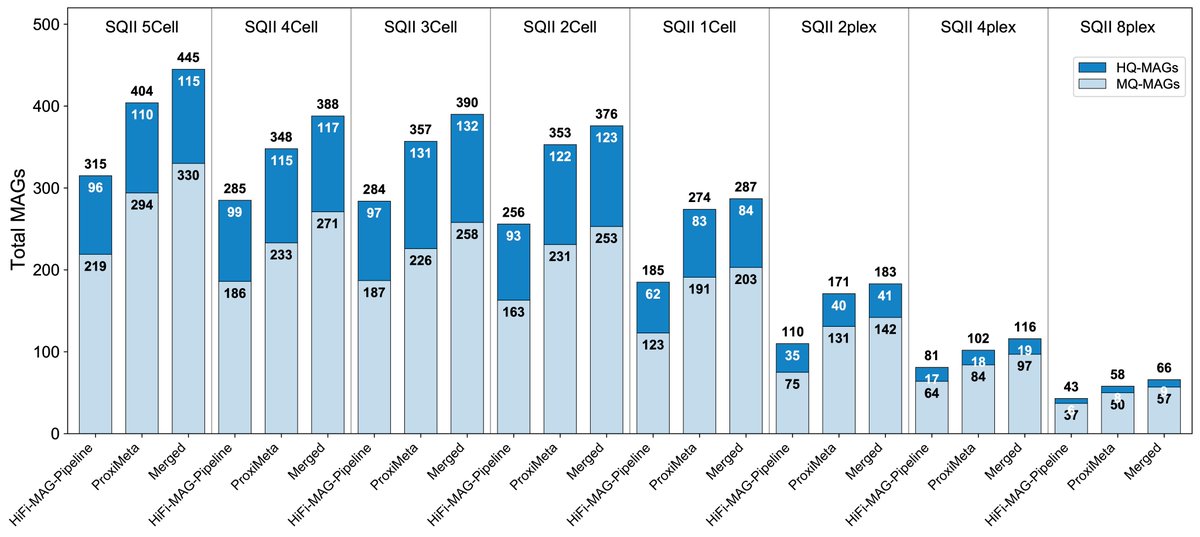
Florian Plaza Oñate Luis Pedro Coelho🌻🇮🇱 Big Data Biology Lab I also wanted to show the pattern is true even for downsampled data. The previous figure shows ~17 million reads (~120Gb) for Revio and SQII, and the combined dataset is ~255Gb.
The figure shown here ranges from 102Gb to 3Gb, and still Hi-C is recovering more total and HQ-MAGs.

AI/ML tools are revolutionising biomedical research as well.
Latest in npj Journals Systems Biology & Applications journal: how to use active learning to optimise cell culture medium for mammalian cells?
nature.com/articles/s4154…
Centre for Predictive Human Model Systems Biodock AI Big Data Biology Lab Manav - The Human Atlas Initiative

Machine learning helps predict prokaryotic antimicrobial peptides using metagenomic and microbial genome datasets.
Preprint from the labs of César de la Fuente and Big Data Biology Lab, evaluated by ArcadiaScience.
sciety.org/articles/activ…

Any plans to use Hi-C interactions to create must-link constraints in SemiBin ? Big Data Biology Lab

I am glad to be joining Big Data Biology Lab during spring as remote intern and am looking forward to learn and contributes during the internship period.
#bioinformatics #remote #intern #skills
BDB-Lab Spring 2023 Updates, by Big Data Biology Lab open.substack.com/pub/bigdatabio…

Second talk for today at #MVIF .28
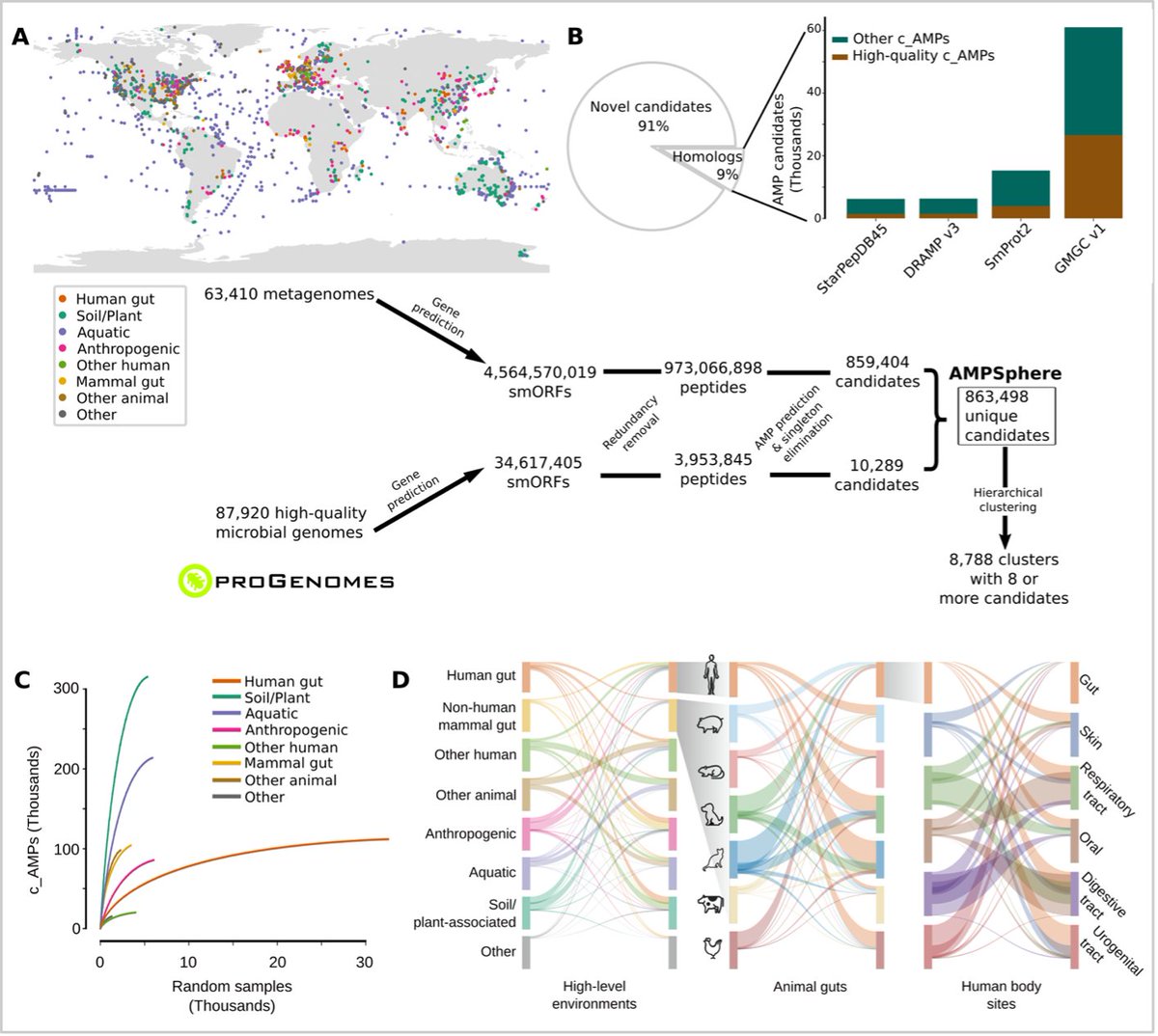
⭐️ A catalogue of small proteins from the global microbiome
by 🇨🇳 Yiqian Duan Yiqian Duan / Big Data Biology Lab
x.com/bigdatabiology…

Luis Pedro Coelho🌻🇮🇱 Svetlana U. Perović ARG annotation in metagenomes is still not solved! ... Nevertheless, the work from Big Data Biology Lab, lead by Svetlana U. Perović and Luis Pedro Coelho🌻🇮🇱 is working to solve it!





SemiBin2 works extremely well for #metagenomic contig binning, and is one of the only methods that currently has long-read specific settings.

Luis Pedro Coelho🌻🇮🇱 Svetlana U. Perović Big Data Biology Lab @Seq4AMR We would like to thank everyone who joined us today for the #EMBARK2023 webinar, our speakers and moderator!
You are welcome to join us for the next one on June 14th with Frank van Leth🇪🇺 FrankVanLeth.bsky.social -Project OASIS and Johan BengtssonPalme
Registration link: antimicrobialresistance.eu/category/webin…
