This 3.7 Å structure resolved by #singleparticle #cryoem shows the results of work towards a method for the production of protein #nanomaterials (doi.org/10.1038/s41467…). The data to resolved this structure is available at #EMPIAR right now (ebi.ac.uk/empiar/EMPIAR-…).

DNA is cool and all, but almost everything in bio relies on what DNA does: protein and enzymes. Check out this @NEOdotLIFE article about #QuantumComputing & #ProteinDesign by Ian Haydon. CC IndieBio Seth Bannon 🌻 Ela Madej ❤️🛕 Institute for Protein Design medium.com/neodotlife/qua…

Congrats 🎉🍾Frederikke M on the very successful defense of your Masters thesis on #proteindesign in the Kresten Lindorff-Larsen and Winther labs!


It's late but my bioRxiv with Florian Praetorius is out and I'm too excited not to share it with you all #ProteinDesign : biorxiv.org/content/10.110…

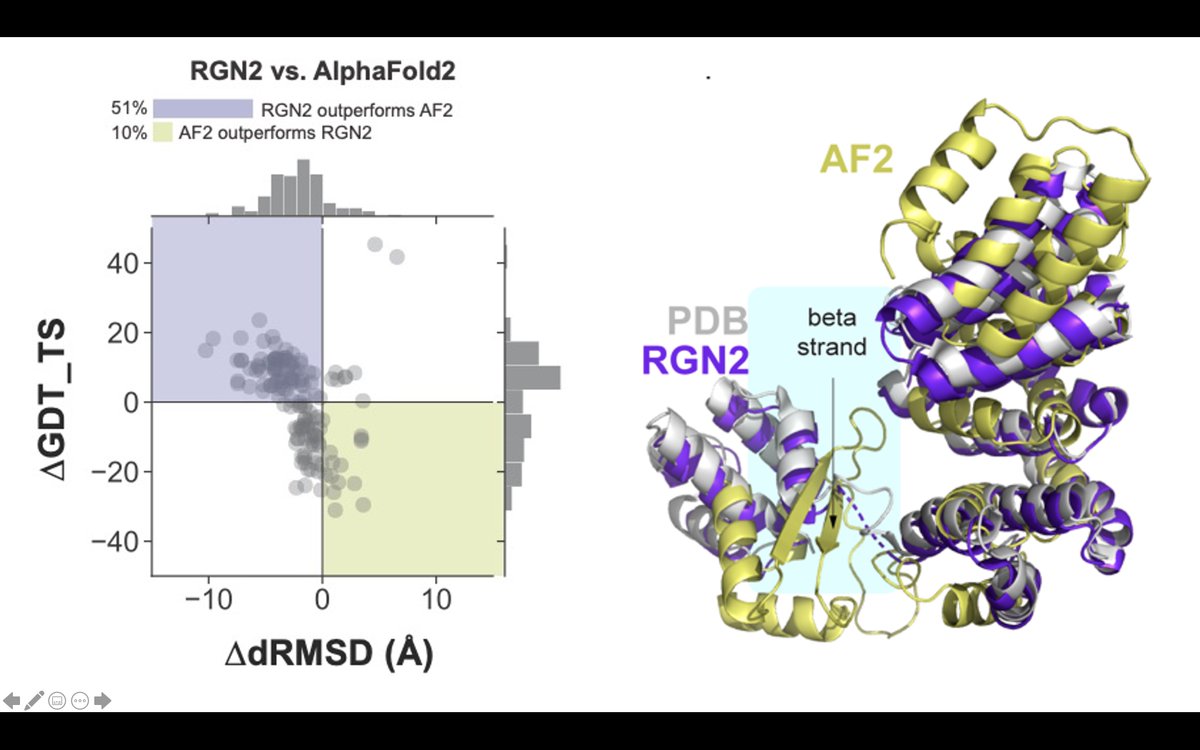
Our RGN2 predictions are several orders of magnitude faster than current methods thus bearing promise for rapid and high-throughput protein library generation and aid #proteindesign
Mohammed AlQuraishi, Peter Sorger, Surge Biswas, Nazim Bouatta,george church


Mark your calendars for the 2021 series of virtual Biocatalysis and Protein Engineering Meetups> Jan 21st (Thur) will feature exciting talks on #DirectedEvolution , #ProteinDesign , ancestral reconstruction, and fluorine #chemistry meetup.com/Virtual-Biocat…

It was my last day working in the amazing WoolfsonLab University of Bristol yesterday. I'm sad to be leaving after six years, but I'm looking forward to starting my Engineering and Physical Sciences Research Council fellowship hosted by @SynthSysEd The University of Edinburgh in a few weeks! #proteindesign

Happy to announce that work from my Masters got published today!
Identification and Analysis of Natural Building Blocks for Evolution-Guided Fragment-Based Protein Design
sciencedirect.com/science/articl…
#ProteinDesign #ProteinEngineering

Hi #ProteinDesign and #AI #DrugDesign enthusiasts, please have a look to openings at Novo in US, UK, and DK and join a great team around Per Greisen Kristine Deibler Che,Yang me and many others! novonordisk.com/careers/career…


Excited to host and listen to the protein-design talk of Prof. David Baker in a special CFEL-seminar on Sep 14th 6 pm CET. Joining forces for XFEL imaging! Open to all!
Institute for Protein Design DESY (English) European XFEL
CFEL CUI: Advanced Imaging of Matter MPI für Struktur und Dynamik der Materie Max Planck Society #proteindesign


Our latest paper on how coiled-coil assemblies tolerate multiple aromatic residues in the hydrophobic core is now published. This work was carried out by Guto Rhys and co-authors @Will_M_Dawson, Joe Beesley, Freddie Martin, Drew Thomson and Leo Brady.
pubs.acs.org/doi/10.1021/ac…

Great collaborative effort by everyone! #ProteinDesign Institute for Protein Design
doi.org/10.1101/2023.0…

We combine a new protein language model (AminoBERT) with an improved version of our end-to-end differentiable machinery (RGN2) to directly generate 3D coordinates. On orphan proteins, RGN2 outperforms all major methods, including #AlphaFold , RoseTTAFold, and trRosetta. (2/4)

I am very exited to give a #FlashPresentation on #Protein_Evolution & #ProteinDesign at the proteinsociety 35th anniversary symposyum this Tuesday 13th of July.
#PS35 #ProteinSociety #conferenceathome #ProteinEngineering #Evolution Paola Laurino
#Cofactors #Coenzymes #Enzymes

#AdvancesInProteinDesign conference has been awesome! Thanks Dror and Julia for organizing this fantastic meeting! 🤩 Don't know about the rabbit ears though 🐰😂
#fascinatedbyenzymes #proteindesign #cranes



Reminder teams: We've organised an awesome seminar this Thursday on #ProteinDesign and Structure & a short update on what's coming up in the challenge! #AlphaFold


📢The registration for the 'Protein Signaling - in space and time'- conference is now open📢
sciencecluster.dk/event/conferen…
#proteomics
#massspectrometry
#proteinsignaling
#proteindesign
#networkbiology
#molecularbiology
#biochemistry
#singlecellanalysis
#novonordiskfoundation

How are buried tyrosine residues tolerated in self-associating peptides? Have a peek Biomacromolecules! DM if you don’t have access to the article. Thanks again to @Will_M_Dawson Joe Beesley Freddie Martin Drew Thomson Leo Brady and Dek Woolfson for your help! #proteindesign #synbio

Wow - it’s my protein! Thanks EMDB - EMPIAR @EBI for the shoutout.
Using the power of computational #proteindesign , we can create proteins with structures and functions never seen before in nature. I created this tetramer using our now-published #Rosetta helical fusion method.
1/3

Roughly one year ago we started out venturing in the world of #proteindesign . Today I am very happy to share our first preprint ‘Efficient and scaleable de-novo protein design using a relaxed sequence space’ (biorxiv.org/content/10.110…) with Sergey Ovchinnikov 🇺🇦 & Hendrik Dietz 1/x

Saw this outdoor ad by the real estate group TaiKooLi in Sanlitun today, don’t know how they came up with this title, but seems they got it: the future of bio.
#evolution #synbio #proteindesign