Mark Budde
@markwbudde
Co-founder and CEO - Plasmidsaurus & Primordium Labs. Prev work: Synthetic mammalian quorum-sensing and pop control; intMEMOIR to optically encode cell lineage.
ID:44451344
http://www.plasmidsaurus.com 03-06-2009 20:39:44
2,4K Tweets
3,4K Followers
2,6K Following

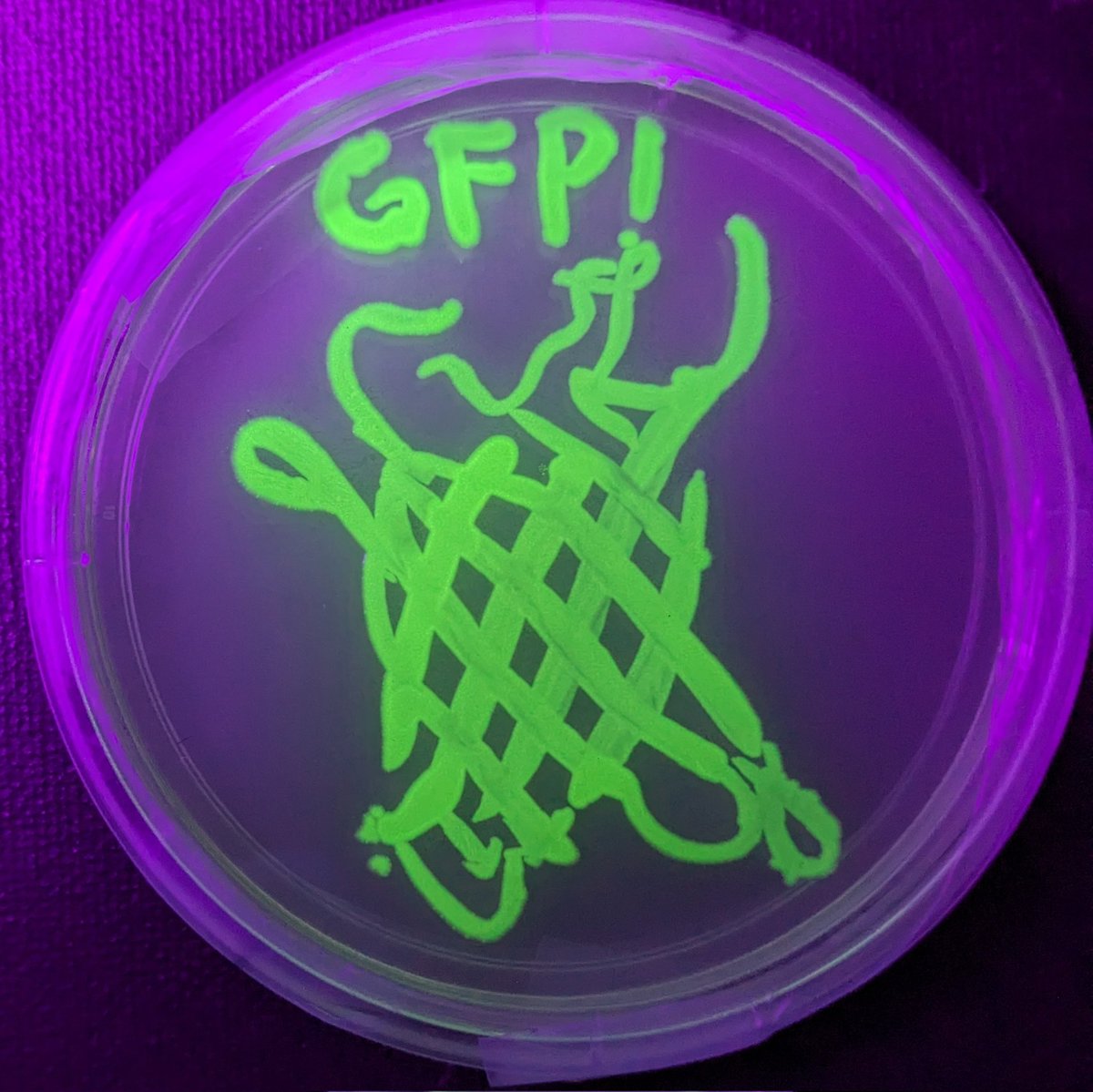
May I present to you my agar art of the structure of green fluorescent protein (GFP), painted with bacteria genetically engineered to express the protein! #meta
Thanks Sebastian S. Cocioba🪄🌷 for the E. coli and Nick Coleman for the super bright free use GFP! #fuGFP #SciArt

Overnight nanopore sequencing service.... roarsome!🦕 #plasmids #microbes #amplicons plasmidsaurus

If you've been using one of our Oxford University dropboxes, you may have spotted something unusual...!!!🦖🚲
Daily pickups at Dunn_School (and @DunnSchool.bsky.social), Oxford Biology, Centre for Human Genetics, and Old Road Campus Research.
Huge shoutout to Dr Malcolm Sim, Malcolm Sim!


Anthony Berndt Kelvin Lau always @klausenhauser on ☁️ 🪡 🐘 Primordium Primordium employee here - hearing that we were able to help made my day :)

Pretty sure we have the fastest and most innovative lab automation team in history at plasmidsaurus 🦖🦕🧬🤖
There is nothing more fun than working with a team of high-agency top performers to unlock new capabilities that others claim are impossible.
This team rocks! 🦾

Thanks to Mark Budde and other alum for hosting a great beer chat! Grateful to have continued connections and proud of his success!


I'm struggling to wrap my head around the new Weissman lab myHSC depletion paper: nature.com/articles/s4158…
The first authors don't seem to be on twitter but hoping I can crowdsource a fun discussion. Daniel Bryan Goodman Michal Tal, PhD Jeffrey Mold Ansu Satpathy Caleb Lareau...





plasmidsaurus I honestly have you guys sequence EVERYTHING before I use it now - even stuff I pull out of my own freezer.
Amazing how much stuff from my own freezer has had duplications...

Anthony Berndt I hate applying it to everything but surely it's an 80:20 rule situation right? 80% of your cloning goes smoothly and only requires 20% of your effort, but that pesky fraction of cloning experiments that don't work the first time are always tough to get working